April 26, 2025 report
This article has been reviewed according to Science X's and . have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Structure of lipid-transfer tunnel protein in C. elegans revealed

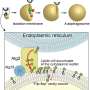
Oregon Health & Science University, in collaboration with Oregon State University, has discovered the structural organization and protein components of a lipid-transfer complex known as LPD-3. Findings show that LPD-3 contains an internal tunnel lined with lipid molecules, suggesting a possible mechanism for large-scale lipid movement between cellular membranes.
Cells must constantly manage the structure and makeup of their membranes, which rely heavily on lipids produced in the endoplasmic reticulum (ER). These lipids cannot freely float through the cytoplasm due to their hydrophobic nature.
Lipid-transport proteins have been shown to shuttle small numbers of lipid molecules between compartments. A distinct group, called bridge-like lipid-transport proteins (BLTPs), may support bulk lipid transfer by forming long, tunnel-like structures that span between organelles. Structural analysis of these proteins has remained limited due to their size and biochemical complexity.
In the study, "Structural basis of lipid transfer by a bridge-like lipid-transfer protein," in Nature, researchers performed cryogenic electron microscopy and mass spectrometry on native LPD-3 isolated from transgenic Caenorhabditis elegans to reveal the subunit composition and structural details of the complex.
Researchers engineered two strains of C. elegans with fluorescent and epitope tags placed at either end of the endogenous lpd-3 gene. The C-terminally tagged strain, which retained normal developmental and cold-stress phenotypes, was used to isolate the native LPD-3 complex.
Proteins were purified from approximately 60 million worms to generate sufficient material for structural and biochemical analysis. Purification was performed using fluorescence-based size-exclusion chromatography. Mass spectrometry identified peptides spanning the full-length 4,022 amino acid LPD-3 sequence.
Structural analysis used single-particle cryogenic electron microscopy to generate a full-length map of the protein at 6.2 angstrom resolution. Functional significance of the complex and its subunits was assessed through RNA interference knockdowns in C. elegans, Drosophila, and HeLa cells.
Cryo-EM analysis resolved only part of the LPD-3 protein, with structural heterogeneity limiting resolution of the C-terminal half.

LPD-3 was resolved as a 345-angstrom elongated tunnel-like structure with a hydrophobic interior filled by 27 lipid molecules and three additional phospholipids within its transmembrane domain. Alternating acidic and basic residues line the tunnel, forming an ionizable track that spans its entire length. Four hydration portals open into the tunnel, permitting cytosolic water access to lipid head groups.
Lipids within the LPD-3 tunnel were spaced approximately 8.4 angstroms apart, a distance that closely resembles their arrangement in natural membrane bilayers.
Mass spectrometry identified two co-purifying auxiliary proteins. One, named Spigot, binds to the N-terminal region of LPD-3 and shares conserved features across species. The second, referred to as lipid transfer auxiliary protein 2 (LTAP2), showed conservation but could not be precisely located in the structural model. A third protein component was also observed as a three-helix transmembrane bundle, but its identity and role remain unclear.
RNA interference knockdown of the spgt-1 gene in C. elegans resulted in a 79.1% reduction in apical actin fluorescence. Knockdown of lpd-3 led to a 91.6% reduction. Knockdown of Spigot orthologs in Drosophila disrupted phagocytosis in astrocytes. Knockdown of C1orf43, the human ortholog of Spigot, in HeLa cells impaired formation of ER–plasma membrane contact sites.
Results establish the subunit composition and molecular architecture of the LPD-3 complex, including how lipid molecules interact with internal tunnel surfaces during transport. Ionizable residues and hydration portals organize lipid head groups along a continuous internal track, suggesting a structural mechanism for bulk lipid transfer.
Findings confirm that Spigot is a conserved component of the complex across multiple species and plays a role in phospholipid transport and cellular organization. Knockdowns of Spigot orthologs in worms, flies, and human cells produced consistent disruptions in membrane-related functions.
The revealed LPD-3 structure provides a foundation for further investigation into BLTP1-related diseases.
Mutations in human BLTP1 are associated with Alkuraya-Kučinskas syndrome, a severe neurological disorder. Structural insights from this study offer a basis for future studies into disease mechanisms and potential therapeutic strategies.
More information: Yunsik Kang et al, Structural basis of lipid transfer by a bridge-like lipid-transfer protein, Nature (2025).
Journal information: Nature
© 2025 Science X Network