This article has been reviewed according to Science X's and . have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Crop phenotyping research: Self-supervised deep learning enhances green fraction estimation in rice and wheat

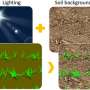
The accurate measurement of the green fraction (GF), a critical photosynthetic trait in crops, typically relies on RGB image analysis employing segmentation algorithms to identify green pixels within the crop. Traditional methods have limitations in accuracy due to environmental variances, while advanced deep learning techniques, like the SegVeg model, show improvement but don't fully leverage the latest vision transformer models.
A significant challenge in applying these state-of-the-art techniques is the lack of comprehensive, annotated datasets for plant phenotyping. Although synthetic image generation offers a partial solution, addressing the realism gap between synthetic and real field images remains a crucial area for future research to enhance the accuracy of GF estimation.
In July 2023, Plant Phenomics published a research article titled " ."
In this study, the objective was to enhance a self-supervised plant phenotyping pipeline for semantic segmentation of RGB images of rice and wheat, considering their contrasting field backgrounds.
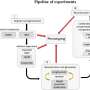
The methodology involved three main steps:
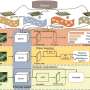
- Collection of real in situ images and their manual annotations from different sites, and generation of simulated images with labels using the Digital Plant Phenotyping Platform (D3P).
- Application of the CycleGAN domain adaptation method to minimize the domain gap between the simulated (sim) and real datasets, creating a simulation-to-reality (sim2real) dataset.
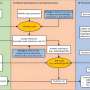
- Evaluation of three deep learning models (U-Net, DeepLabV3+, and SegFormer) trained on real, sim, and sim2real datasets, comparing their performance at pixel and image scales, with a focus on Green Fraction (GF) estimation.
The results showed that domain adaptation through CycleGAN effectively bridged the gap between simulated and real images, as evidenced by improved realism in plant textures and soil backgrounds, and a decrease in Euclidean distance between the sim2real and real images.
The pixel-scale segmentation demonstrated that U-Net and SegFormer outperformed DeepLabV3+, with SegFormer, especially when trained on the sim2real dataset, exhibiting the highest F1-score and accuracy. This trend was consistent for both rice and wheat crops.
The sim2real dataset enabled the best performance in GF estimation, showing close results between simulated and real datasets, especially for wheat. The study also utilized the best-performing model, SegFormer, trained on the sim2real dataset, to explore GF dynamics, effectively capturing the growth stages of rice and wheat, thus indicating accurate GF estimation.
The study also identified critical factors affecting estimation uncertainty, such as nonuniform brightness within images and the presence of senescent leaves. The self-supervised nature of the pipeline, requiring no human labels for training, was emphasized as a significant time-saver in image annotation.
Overall, the research demonstrated that SegFormer trained on the sim2real dataset outperformed other models, highlighting the effectiveness of the self-supervised approach in semantic segmentation for plant phenotyping.
The success of this method opens avenues for further research in enhancing the realism of simulated images and applying more sophisticated domain adaptation models for accurate GF estimation throughout the entire crop growth cycle.
More information: Yangmingrui Gao et al, Enhancing Green Fraction Estimation in Rice and Wheat Crops: A Self-Supervised Deep Learning Semantic Segmentation Approach, Plant Phenomics (2023).
Provided by NanJing Agricultural University