This article has been reviewed according to Science X's and . have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Research unveils new bacteriophage more effective than similar species

Bacteria are becoming increasingly resistant to antibiotics. This "race" can lead to a situation where antibiotics will stop suppressing pathogenic bacteria. Bacteriophages, natural "predators" of bacteria, are considered one of the alternatives.
The Laboratory for Metagenome Analysis at the Skoltech Bio Center works on the research on bacteriophages and bacterial immune systems. In the study, a group of scientists led by the head of the laboratory Artem Isaev analyzed the composition of one of the therapeutic phage cocktails and discovered a new type of bacteriophage—it turned out to be more effective than previously known similar species.
The are published in Viruses.
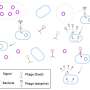
Phage cocktails containing a mixture of phages selected from natural sources are available on the pharmaceutical market in some countries, including Russia. Often, the composition of these mixtures is not disclosed, so phages remain unexplored at the genomic level. The team analyzed the Sextaphage cocktail from the Russian company Microgen—a national manufacturer of immunobiological drugs.
"We conducted a study on laboratory E. coli bacteria. We selected phages that can infect it, sequenced their genomic DNA, and found an interesting Sxt1 phage," said Polina Iarema, the lead author of the work, a Ph.D. student of the Life Sciences program.
"It can infect not only laboratory E. coli, which is convenient to work with because it doesn't have protective systems, but also some natural isolates that are resistant to most phages due to the intricacies of the cell wall structure. This surprised us, as the closest relatives of Sxt1 are not able to overcome these barriers. We assume that it will handle pathogenic infections more effectively."
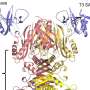
The results showed that the Sxt1 genome is extremely similar to the genomes of T3 and T7 phages of the Autographiviridae family. Their distinctive feature of this family is viral RNA polymerase, which transcribes the phage genome.
"Despite the fact that Sxt1 does not differ very much in the genomic sequence, it infects bacteria in a different way. Our investigation began by examining the genes encoded in this bacteriophage and how they differ from those of existing bacterial phages, in order to understand the exact cause of its broader specificity. We found that this bacteriophage has different fibrils—or 'legs'—and receptors on them that recognize bacteria," added Oksana Kotovskaya, a co-author of the work and a Ph.D. student of the Life Sciences program.
A group of researchers has taken an important step towards a detailed study of the composition of phage cocktails. An accurate understanding of their components will help make sure they are safe for humans.
More information: Polina Iarema et al, Sxt1, Isolated from a Therapeutic Phage Cocktail, Is a Broader Host Range Relative of the Phage T3, Viruses (2024).
Provided by Skolkovo Institute of Science and Technology