This article has been reviewed according to Science X's and . have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Researchers publish gapless genome assembly for East Asian finless porpoise

The East Asian finless porpoise (Neophocaena asiaeorientalis sunameri) is a small, freshwater cetacean species. This species is a group of small-sized, toothed whales that are mainly distributed in southern and eastern Asia. Their distribution includes the coastal waters of the western Pacific Ocean, Indian Ocean, Sea of Japan, and they also appear in the Bohai Sea, Yellow Sea, East China Sea, South China Sea, and middle and lower reaches of the Yangtze River in Chinese waters. Previous research found that the genetic differentiation between the Yangtze finless porpoise (N. a. asiaeorientalis) and East Asian finless porpoise reached interspecific level.
In this study, published in the journal Scientific Data, researchers from BGI Genomics helped generate a gapless genome assembly for the East Asian finless porpoise to provide new resources for the comparative genomics of cetaceans and conservation biology of threatened species.
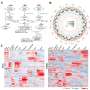
A gapless genome assembly means that the entire genome has been pieced together without any gaps or missing sections. Generating a gapless genome assembly is a significant achievement because it allows for a more accurate understanding of an organism's genetics and can provide insight into its evolutionary history.
The researchers succeeded in generating a gapless genome assembly for the finless porpoise, which is a significant achievement. It approximately is 2.5 Gb in size over its 21 autosomes and two sex chromosomes. A total of 22,814 protein-coding genes were predicted where 97.31% were functionally annotated, which means that 95.3% of the genome has been pieced together into continuous stretches without any gaps.
The genome assembly also has a scaffold N50 of 1.2 Mb, which is a measure of the size of the largest contiguous stretches of the genome. A larger scaffold N50 indicates a higher quality whole genome DNA sequence reconstruction. The researchers also identified a huge quantity of protein-coding genes in the genome assembly.
In addition to generating the genome assembly, researchers also performed various analysis to investigate the finless porpoise's evolutionary history and adaptation to its aquatic environment. They found out that this cute cetacean has genetic adaptations that are likely important for its survival in freshwater habitats, including genes involved in osmoregulation (the regulation of water and electrolyte balance in the body) and detoxification.
The researchers also identified a number of genes involved in hearing, olfaction, and immunity that are unique to the finless porpoise.
Overall, this study represents an important step forward in our understanding of the East Asian finless porpoise and its genetics. The gapless genome assembly and identification of unique genes will likely provide a valuable resource for future research on this species and its evolutionary history.
In the future, this research may help us to better understand and protect this animal, which is facing numerous threats in its natural habitat. It may also be useful for comparative studies with other cetacean species and for understanding the evolutionary history of cetaceans as a whole.
More information: Denghua Yin et al, Gapless genome assembly of East Asian finless porpoise, Scientific Data (2022).
Journal information: Scientific Data
Provided by BGI Genomics