New study increases understanding of how antibiotic resistance arises
How does antibiotic resistance arise? In a new article in the July number of the Journal of Biological Chemistry researchers from Uppsala University show how a bacterial enzyme can learn to inactivate antibiotic molecules by mutating.
Bacteria can use many different methods to outwit the antibiotic molecules we use to treat communicable diseases. One common method is for a bacterial enzyme to inactivate the antibiotic molecule by modifying it chemically.
"This means that the antibiotic molecule no longer fits its binding position and cannot exert its effect – in the same way that a bandaged foot no longer fits its shoe," explains Maria Selmer.
One reason why bacteria can quickly become resistant to new antibiotics is that enzymes can learn to inactivate several different antibiotic molecules by mutating.
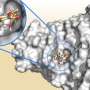
In a new article in the July number of the Journal of Biological Chemistry Maria Selmer's research group shows how an antibiotic resistance enzyme (AadA) is able to recognise and inactivate two chemically different antibiotics (streptomycin and spectinomycin). Using crystallography (a method for determining protein structure), computer simulations and biochemistry the researchers show at atomic level how different antibiotic molecules fit into different parts of the bacteria enzyme where the inactivation reaction occurs.
"This detailed knowledge about how the enzyme recognises two different antibiotics can help us to understand how an existing resistance enzyme can be upgraded and inactivate additional types of antibiotics, leading to a rapid development of resistance to new antibiotics," says Selmer.
More information: Ana Laura Stern et al. Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica, Journal of Biological Chemistry (2018).
Journal information: Journal of Biological Chemistry
Provided by Uppsala University