Scientists one step closer to cracking the mystery of bacterial adaptation to antibiotics
An international team including researchers from MIPT's Laboratory for Advanced Studies of Membrane Proteins have proposed an explanation of how bacteria process external signals. By identifying the detailed structure of the protein complex used by bacteria, the scientists gained insights into the ability of these microorganisms to detect even small changes in the environment and adapt to them. The research findings were published in Scientific Reports.
Bacteria are extremely good at adapting to a changing environment. This renders many antibiotics ineffective, as the bacterial cell can adapt by developing resistance. Because resistant bacteria can survive the influence of drugs, infectious diseases can become difficult to treat.
To gather information about the outside world, bacteria rely on two-component signal systems constituted by transmembrane protein complexes, i.e., structures made up of two proteins in the cell membrane, one of them "sticking out" and the other extruded inward. To understand the mechanism behind the operation of such complexes, researchers need to determine their precise structure. Knowing how this system works, scientists could then figure out how to switch it off. This makes membrane protein complexes potentially useful targets for emerging antibiotics.
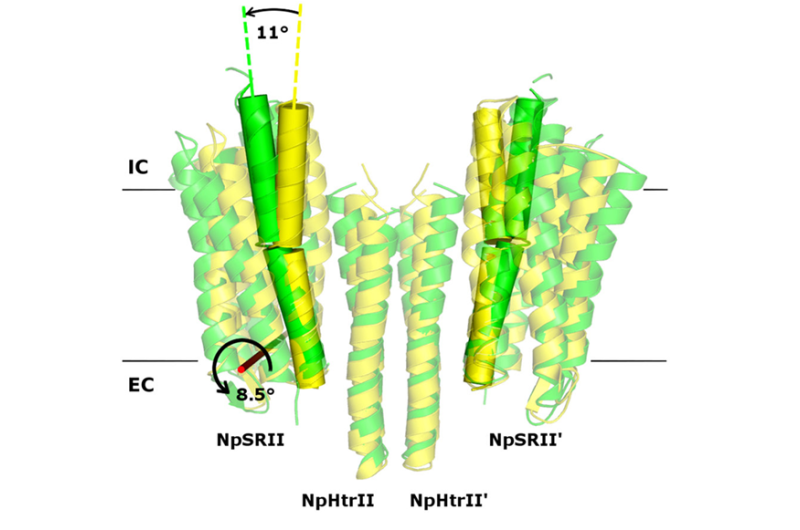
In their study, the researchers examined the crystal structures of the ground and active states of one such system consisting of the sensory photoreceptor rhodopsin II coupled with its cognate transducer. The team has demonstrated that this complex can have a U-shaped structure in addition to the regular V-shaped conformation reported in prior research. The study also explains why this is the case by positing biological relevance of the U-shape in terms of signal transduction.
The team suggested that by transitioning from the U- to the V-shape, the receptor–transducer complex enters its active state, which could be involved in signal transmission between the photoreceptor and the transducer. This is in line with existing biological data. Therefore, it is possible to disrupt signal transduction by treating the cell with a suitable drug preventing the V-to-U transition.

"Our findings have a practical application in dealing with bacterial resistance. Nevertheless, this study is primarily significant for our fundamental understanding of signal transduction mechanisms in bacteria, because they could be involved in thousands of other similar bacterial receptors that are responsible for all kinds of cell functions. These insights will enable us to come up with receptor models that are more precise," says Valentin Borshchevskiy, a senior research scientist at the Laboratory for Advanced Studies of Membrane Proteins.
Among other bacterial receptors, the protein complex investigated in the study plays a major role in the aspartate (Tar) and serine (Tsr) receptors. The former guides bacteria toward nutrients (e.g., aspartate and maltose) and away from harmful agents (e.g., nickel and cobalt). The latter is used by bacteria like Salmonella and E. Coli to seek serine, which they consume as a nutrient, and avoid harmful acids.
More information: A. Ishchenko et al. New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex, Scientific Reports (2017).
Journal information: Scientific Reports
Provided by Moscow Institute of Â鶹ÒùÔºics and Technology